New Open-Source Tool Could Speed Up Diagnoses
for Children with Rare Diseases
For families of children with rare diseases, the road to finding a diagnosis can be long and heartbreaking. Parents often face months or even years of visiting specialists, undergoing countless tests, and waiting for results—all while their child’s condition remains a mystery.
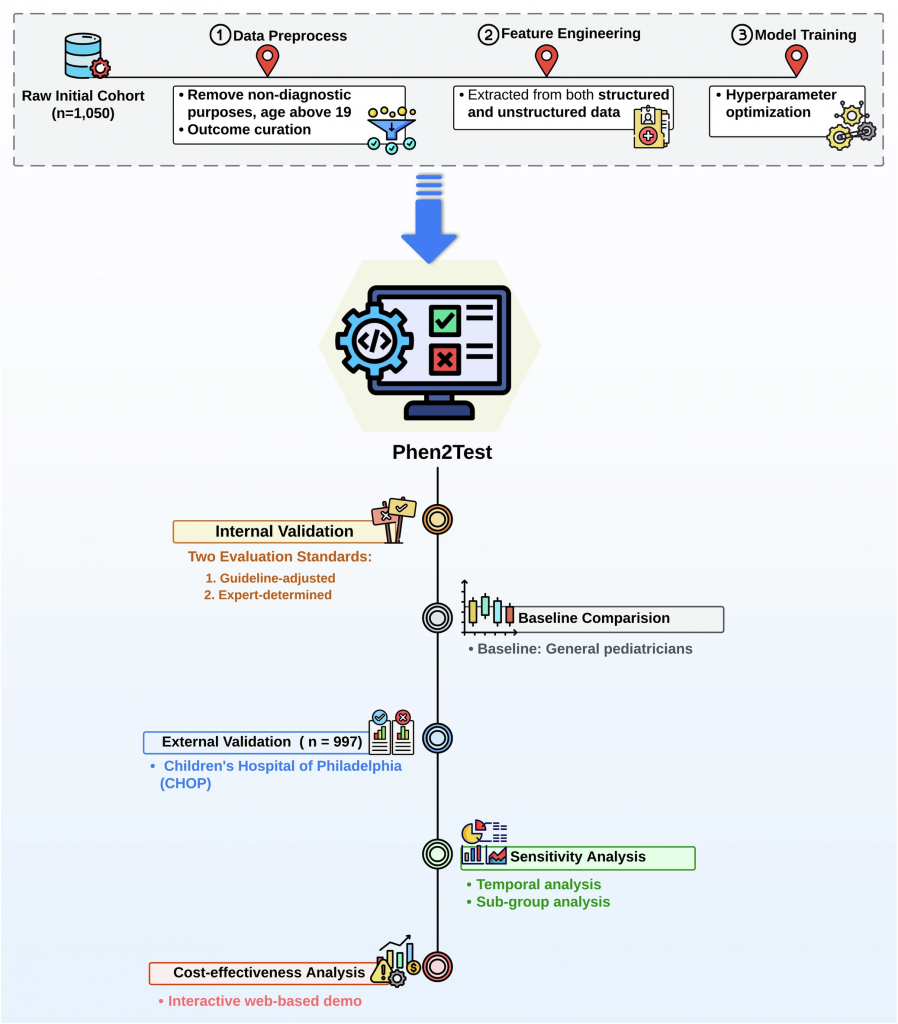
Researchers in Dr. Chunhua Weng’s Lab at the Columbia University Vagelos College of Physicians and Surgeons (VP&S) are working on a solution to make this process faster and more efficient. Drs. Liu and Weng and Ms. Fangyi Chen have developed a model called Phen2Test that uses artificial intelligence (AI) to recommend the most appropriate and needed genetic test for children who might have rare diseases to primary care providers.
“Selecting the appropriate genetic test as early as possible can make a huge difference for families, both clinically and socioeconomically” says Chen, a current PhD student in Biomedical Informatics at Columbia University. “It can avoid unnecessary costly tests, save time for healthcare providers and families, reduce stress, and identify the right treatment sooner.”
Phen2Test is a novel machine learning model that analyzes multimodal data from a child’s electronic medical records—such as symptoms and other health details—and recommends the most appropriate genetic test, such as sequencing specific genes or scanning the entire genome.
“There is a severe shortage of genetic counselors across the country, delaying the diagnosis of genetic disorders,” Chen said. “In essence, Phen2Test serves as a portable digital genetic counselor, guiding the selection of genetic tests at the point of care.”
This tool can be especially useful to primary care providers who aren’t genetic specialists and don’t have the expertise to select the right tests on their own. Phen2Test bridges that gap, offering expert-level support to primary care doctors and pediatricians. It also stays up-to-date with the latest clinical guidelines, so its recommendations are current and reliable.
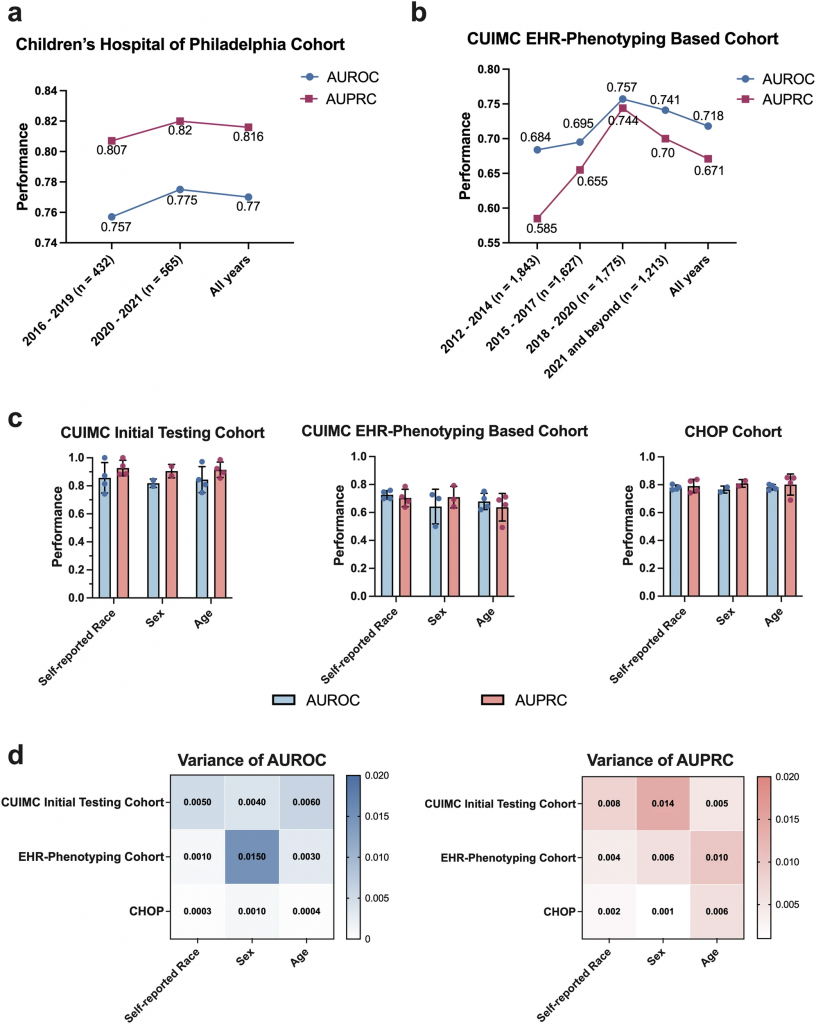
The tool was tested on more than 1,000 pediatric patients at Columbia University’s hospital and was later validated at the Children’s Hospital of Philadelphia (CHOP). In both cases, it performed remarkably well at choosing the same tests that genetic specialists would pick (see figure below).
The researchers are looking for ways to enhance the tool, including how it could incorporate facial photos, which are commonly available in electronic health records systems for recognizing patients, to provide richer phenotypes. Financial costs and resource availability are relevant practical factors in the real-world decision-making process, but they are not currently accounted for within the tool. Therefore, future work is warranted to incorporate practical factors into this model using insurance data together with clinical data.
Creating trust in AI solutions among clinicians also remains a healthcare obstacle for AI interventions.
“While Phen2Test performs comparably to genetic specialists, general pediatricians may lack confidence in using it,” Chen said. “Building trust around AI and tools like Phen2Test is a sociotechnical challenge. We need to make sure doctors feel comfortable using it. We also need to conduct more studies to further ensure the safety and robustness of the tool.”
The study, recently published by Nature Digital Medicine, shows that Phen2Test has the potential to make a real difference for children and their families. The researchers have made both the code and trained model for Phen2Test publicly available on GitHub.
Additional Information
The study, “Phenotype driven molecular genetic test recommendation for diagnosing pediatric rare disorders,” was published Nov. 21 in NPJ Digital Medicine.
All authors (from Columbia unless noted): Fangyi Chen, Priyanka Ahimaz, Quan M. Nguyen (University of Pennnsylvania), Rachel Lewis, Wendy K. Chung (Harvard Medical School), Casey N. Ta, Katherine M. Szigety (University of Pennnsylvania), Sarah E. Sheppard (University of Pennnsylvania), Ian M. Campbell (University of Pennnsylvania), Kai Wang, Chunhua Weng & Cong Liu (Harvard Medical School).
This study was supported by National Human Genome Research Institute grant R01HG012655, and National Center for Advancing Translational Sciences, National Institutes of Health (NIH), through Grant Number UL1TR001873.