DBMI Research
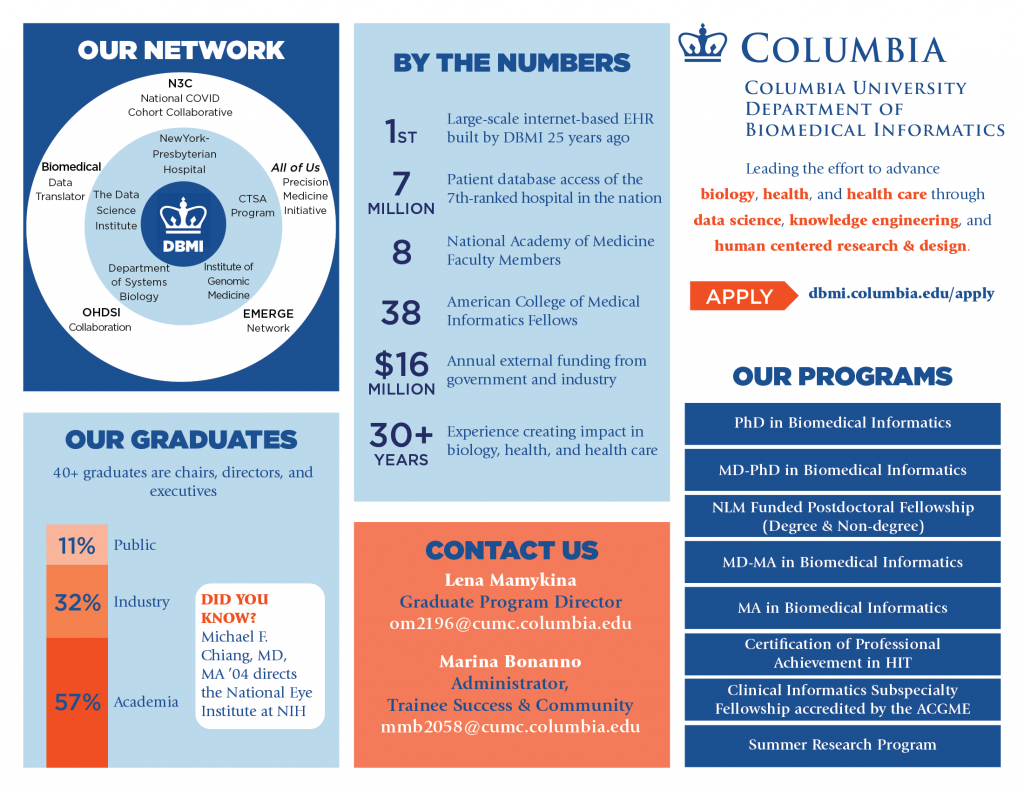
The research mission within the Columbia University Department of Biomedical Informatics is to produce theoretical and practical advances in AI, data science, and human-computer interaction, which will lead to better health and new biomedical knowledge.
DBMI faculty and trainees work collaboratively to design, implement, and evaluate innovative informatics-driven solutions to enable clinicians, patients & communities, and researchers. We also develop, validate, and deploy novel computational methodologies to advance research in AI and in health.
One of the oldest biomedical informatics departments in the nation, DBMI has brought together leaders from a wide range of specialties to lead novel initiatives and train the next generation of researchers. Learn about our research advisors and their respective focuses.